Coordinate Descent for Logistic Regression
A walk-through of two coordinate descent variants on cross-entropy loss for a binary UCI wine task, compared against scikit-learn's logistic regression baseline. Coordinate descent is a fundamental optimization tool in machine learning because it scales to high-dimensional problems, lets us update one parameter at a time, and underpins many modern large-scale training routines.
Introduction
This post showcases two different approaches to implementing coordinate descent on a model defined with cross-entropy loss. Coordinate descent can struggle with non-smooth multivariate functions, as the algorithm may get stuck at non-stationary points where level curves are not smooth. However, it has gained significant interest for large-scale problems, where it has proven competitive with other algorithms for training support vector machines and performing non-negative matrix factorization.
In this experiment I implement coordinate descent using the following approaches:
- Randomly optimize along a random coordinate
- Optimize along coordinate with the largest gradient in magnitude
To benchmark the solutions I implemented a standard logistic regression model without focusing on validation and was just used to obtain scikit-learn's implementation of gradient descent and final loss. The model is trained with the wine.data dataset provided by UCI machine learning repository.
Procedure
I use the cross-entropy loss \(L(w)\) in the coordinate descent method. Let \(\hat{y}\) be the probability of having label 1, and let \(y\) be the actual label. Then the cross-entropy loss and gradient is defined below:
\[ L(w) = -\frac{1}{n}\sum_{i=1}^{n} y_{i} \log \hat{y}_{i} + (1 - y_{i}) \log(1 - \hat{y}_{i}) \]
And thus the gradients with respect to each weight \(w_{j}\) is given by:
\[ \frac{\partial L(w)}{\partial w_{j}} = \sum_{i=1}^{n}(\hat{y}_{i} - y_{i})x_{j} \]
The loss function \(L(\bullet)\) should be differentiable and preferably convex. The procedure of the coordinate descent method is outlined below:
- Initialize weights to zero
- Get predictions from sigmoid function
- Calculate loss and gradients
- Find coordinate \(i\) with the largest gradient in absolute value (best coordinate descent) or randomly choose weight (random coordinate descent)
- Update weight \(i\) via: \(w_{i} = w_{i} - \eta \frac{\partial L(w)}{\partial w_{i}}\)
- Repeat steps until convergence
Data Preparation
import numpy as np
from tqdm import tqdm
from random import randint
import matplotlib.pyplot as plt
from sklearn.metrics import log_loss
from sklearn.metrics import accuracy_score
from sklearn.linear_model import LogisticRegression
data = np.loadtxt('./data/wine.data', delimiter=',')
X, y = data[:, 1:], data[:, 0]
# transform problem into binary classification task
idxs = [i for i in range(len(y)) if y[i] == 1 or y[i] == 2]
X, y = X[idxs], y[idxs]
# normalize data
X = (X - X.mean(axis=0))/(X.max(axis=0) - X.min(axis=0))
X = np.hstack((X,np.ones(len(X)).reshape(len(X),1)))
# transform target variable
y = np.array(list(map(lambda x: 0 if x == 1 else 1, y)))
Now that the data is prepared, we can use scikit-learn's implementation as our benchmark:
# scikit learn implementation (our benchmark)
reg = LogisticRegression(solver='sag', C=100000, max_iter=10000).fit(X,y)
L_star = log_loss(y,reg.predict_proba(X))
print("Loss L* = {:<16f}".format(log_loss(y,reg.predict_proba(X))))
Loss \(L^{*} = 0.000393\). We can now benchmark our coordinate descent implementations against this baseline \(L^{*} \approx 0.0004\). First let's define a few helper functions:
def sigmoid(x):
return 1 / (1 + np.exp(-x))
def cross_entropy_loss(y,y_pred):
loss = -(1/len(y))*np.sum(y*np.log(y_pred) + (1 - y)*np.log(1 - y_pred))
return loss
def cross_entropy_grad(y,y_pred,x):
return list(np.dot((y_pred - y),X)[0])
Best Index
For each iteration, only update weight \(w_{j}\) such that \(|\frac{\partial L(w)}{\partial w_{j}}|\) is the largest for all \(j\).
# initial weights
w = np.zeros(14).reshape(14,1)
w_rand = w
eta = 0.01
loss = []
loss_rand = []
num_iter = 1005000 # just take a bunch of iterations
for t in tqdm(range(num_iter)):
# predict step
y_pred = sigmoid(np.dot(w.T,X.T))
grad = cross_entropy_grad(y,y_pred,X)
largest = np.argmax(np.abs(grad)) # idx with largest magnitude
loss.append(cross_entropy_loss(y,y_pred))
# update that coordinate with largest gradient in magnitude
w[largest] = w[largest] - eta*grad[largest]
y_pred = sigmoid(np.dot(w.T,X.T))
y_pred = np.array(list(map(lambda x: 1 if x >= 0.5 else 0, y_pred.flatten())))
print("Accuracy for best coordinate descent {0}".format(accuracy_score(y, y_pred)))
print("Loss L = {0}".format(loss[-1]))
Accuracy for best coordinate descent 1.0
Loss = 0.0004698390523920375
Random Index
For each iteration, update weight \(w_{j}\) for \(j \in 0 \dots 13\) (e.g., randomly choose a coordinate to minimize on).
# initial weights
w = np.zeros(14).reshape(14,1)
w_rand = w
eta = 0.01
loss_rand = []
num_iter = 1005000
for t in tqdm(range(num_iter)):
# predict step
y_pred = sigmoid(np.dot(w.T,X.T))
loss_rand.append(cross_entropy_loss(y,y_pred))
grad = cross_entropy_grad(y,y_pred,X)
random = randint(0,13)
# update random coordinate
w_rand[random] = w_rand[random] - eta*grad[random]
y_pred = sigmoid(np.dot(w_rand.T,X.T))
y_pred = np.array(list(map(lambda x: 1 if x >= 0.5 else 0, y_pred.flatten())))
print("Accuracy for random coordinate descent {0}".format(accuracy_score(y, y_pred)))
Accuracy for random coordinate descent 1.0
plt.figure(figsize=(10,10))
plt.plot(loss, 'g-', label='Best Coordinate Descent')
plt.plot(loss_rand, 'r-', label='Random Feature Coordinate Descent')
plt.axhline(y=L_star, color='y', label='L*')
plt.title('iteration vs loss', fontsize=25)
plt.xlabel('iteration', fontsize=15)
plt.xlim(0,1000000)
plt.ylim((-0.01,0.04))
plt.ylabel('loss', fontsize=15)
plt.xticks(fontsize=15)
plt.yticks(fontsize=15)
plt.grid()
plt.legend()
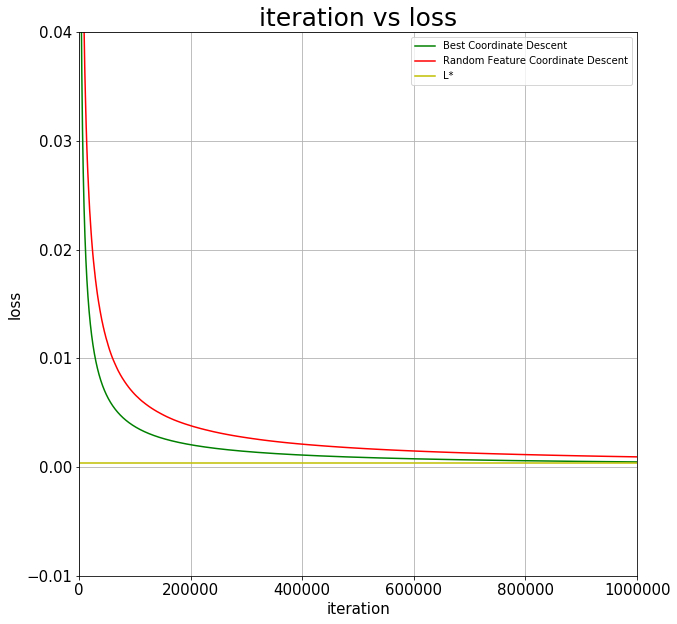
Experimental Results
In this experiment I began by running an unregularized standard logistic regression solver from scikit-learn. The final cross-entropy of the logistic regression model is \(L^{*} \approx 0.0004\).
In my results, after 1.5 million updates, the coordinate descent method yields a loss \(L = 0.00047\), showing convergence to \(L^{*}\). It is important to note that both implementations of coordinate descent (random feature selection and best feature selection) converge to the loss \(L^{*}\), but at different rates.
We conclude that coordinate descent is more optimally implemented if the coordinate chosen has the largest gradient in magnitude. A clear advantage can be seen above from randomly choosing coordinates to update on.